Having changed approaches a lot (studying biochemistry, chemistry, computational biology and most recently theoretical/modeling/computational work), I am very aware that specific fields use specific jargon to describe their work. I tried to make these summaries understandable to different fields, but some jargon and field specific vocabulary remains (especially in the chemistry section). If you want to read a more 'general public' view of my past and current research, head over to this article I wrote for Biomusings: "My astrobiologist view of life".
You can also get an idea of my more recent work on the podcast episode I did on SFI's complexity podcast.
Finally, I was involved in a collaborative writing project involving many early career researchers, meant to help introduce the field of Astrobiology for students and other scientists wishing to gain knowledge in this growing field by crafting one handy collection: the Astrobiology primer 3.0. I was involved as chapter editor and writer on two chapters: Chapter 8: Searching for Life Beyond Earth, and Chapter 9: Life as We Don't Know It.
I am particularly proud to have co-conceived the later chapter (along with Lu Chou) as it is a subject that I believe is of vital importance when considering the search of life beyond Earth.
You can also get an idea of my more recent work on the podcast episode I did on SFI's complexity podcast.
Finally, I was involved in a collaborative writing project involving many early career researchers, meant to help introduce the field of Astrobiology for students and other scientists wishing to gain knowledge in this growing field by crafting one handy collection: the Astrobiology primer 3.0. I was involved as chapter editor and writer on two chapters: Chapter 8: Searching for Life Beyond Earth, and Chapter 9: Life as We Don't Know It.
I am particularly proud to have co-conceived the later chapter (along with Lu Chou) as it is a subject that I believe is of vital importance when considering the search of life beyond Earth.
Research Interests
I am interested in developing a deeper understanding of the organizing principles of biology and its very nature, and gain better understanding of life’s emergence on Earth and elsewhere.
I have always been interested in understanding life, how it started on Earth or could start in different environments, and what makes it different to 'non-life'. This fascination with understanding life in the universe compelled me to study biochemistry at University College London (UCL) so that I might better understand how life, at least as it is here on Earth, works. I later went on to do my Ph.D. at UCL in prebiotic chemistry in order to help uncover the chemical processes potentially present at the start of life on Earth. I then did a postdoc at the Santa Fe Institute, working with Chris Kempes on the Agnostic Biosignatures project - a multi-institution NASA-funded project interested in understanding how to search for life in the universe without presupposing a particular biochemical framework. During this postdoc I worked on projects related to understanding what patterns or signatures life might generate universally by working towards understanding general principles of life; as well as how to detect proposed agnostic biosignatures using current technology.
I have always been interested in understanding life, how it started on Earth or could start in different environments, and what makes it different to 'non-life'. This fascination with understanding life in the universe compelled me to study biochemistry at University College London (UCL) so that I might better understand how life, at least as it is here on Earth, works. I later went on to do my Ph.D. at UCL in prebiotic chemistry in order to help uncover the chemical processes potentially present at the start of life on Earth. I then did a postdoc at the Santa Fe Institute, working with Chris Kempes on the Agnostic Biosignatures project - a multi-institution NASA-funded project interested in understanding how to search for life in the universe without presupposing a particular biochemical framework. During this postdoc I worked on projects related to understanding what patterns or signatures life might generate universally by working towards understanding general principles of life; as well as how to detect proposed agnostic biosignatures using current technology.
Past research
Agnostic biosignatures
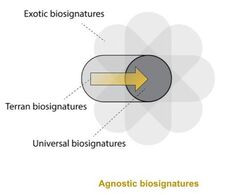
During my postdoc I worked on several projects that are generally interested in understanding how we can look for life when we don't know how different life could be to Terran life (i.e. known life on Earth). This involves proposing new "agnostic biosignatures", or signatures of life that don't presuppose what biochemistry other life forms would be composed of. In this vain I am currently involved in several projects that are interesting in understanding how we could use tools at our disposal (e.g. mass spectrometers) to look for life in the general sense; as well as understanding how certain dynamics, like evolution, can be linked to distinctive patterns in (theoretical) molecular systems.
You can find my recent white paper (written with Laboratory of Agnostic Biosignatures colleagues for the Planetary Science and Astrobiology Decadal Survey 2023-2032) here, which makes the case for a more agnostic approach to life detection, and describes a few new approaches in this field of research.
You can find my recent white paper (written with Laboratory of Agnostic Biosignatures colleagues for the Planetary Science and Astrobiology Decadal Survey 2023-2032) here, which makes the case for a more agnostic approach to life detection, and describes a few new approaches in this field of research.
Prebiotic chemistry and the origin of life
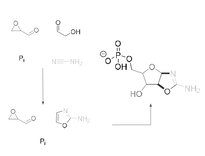
I did my PhD at University College London with Matthew Powner, and in our lab, one of the main interests was in understanding how the building blocks of ribonucleic acid (RNA) could be assembled from simple molecules that are thought to have been present on the early Earth. My PhD was focused on studying a prebiotically plausible way of making phosphorylated ribonucleotides and amino acids. My main project, which was compiled into two publications, focused on getting closer to a one-pot reaction making pentose aminooxazolines, one of the key intermediates in the synthesis of ribonucleotides, by solving the need for spatial and temporal separation of the two and three carbon sugars (feedstock molecules) and finding a prebiotically plausible way of incorporating phosphates into these intermediates while working in water. I also worked on integrating this work with amino acid and other metabolite synthesis, in line with a systems chemistry approach to the origin of life.
Canonical amino acids and the origin of their use in biology
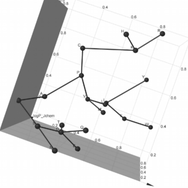
After my PhD, I had the chance to work with Jim Cleaves at the Earth Life Science Institute in Tokyo, doing a short computational biology project there with a large international team. The project built upon a previous publication aimed at understanding why we use the 20 amino acids (canonical amino acids) that we do in biology, when there are many other options out there. They had looked at how well our 20 amino acids covered the chemical space (here defined as charge, hydrophobicity and mass ranges) compared to some other 20 amino acids (selected at random from a large pool of possible amino acids). In this project, we explored how well subsets of those 20 amino acids vs smaller sets of random amino acids cover the chemical space, and found that they did so particularly well, giving those canonical sets an adaptive advantage.
